Centromere organization and evolution
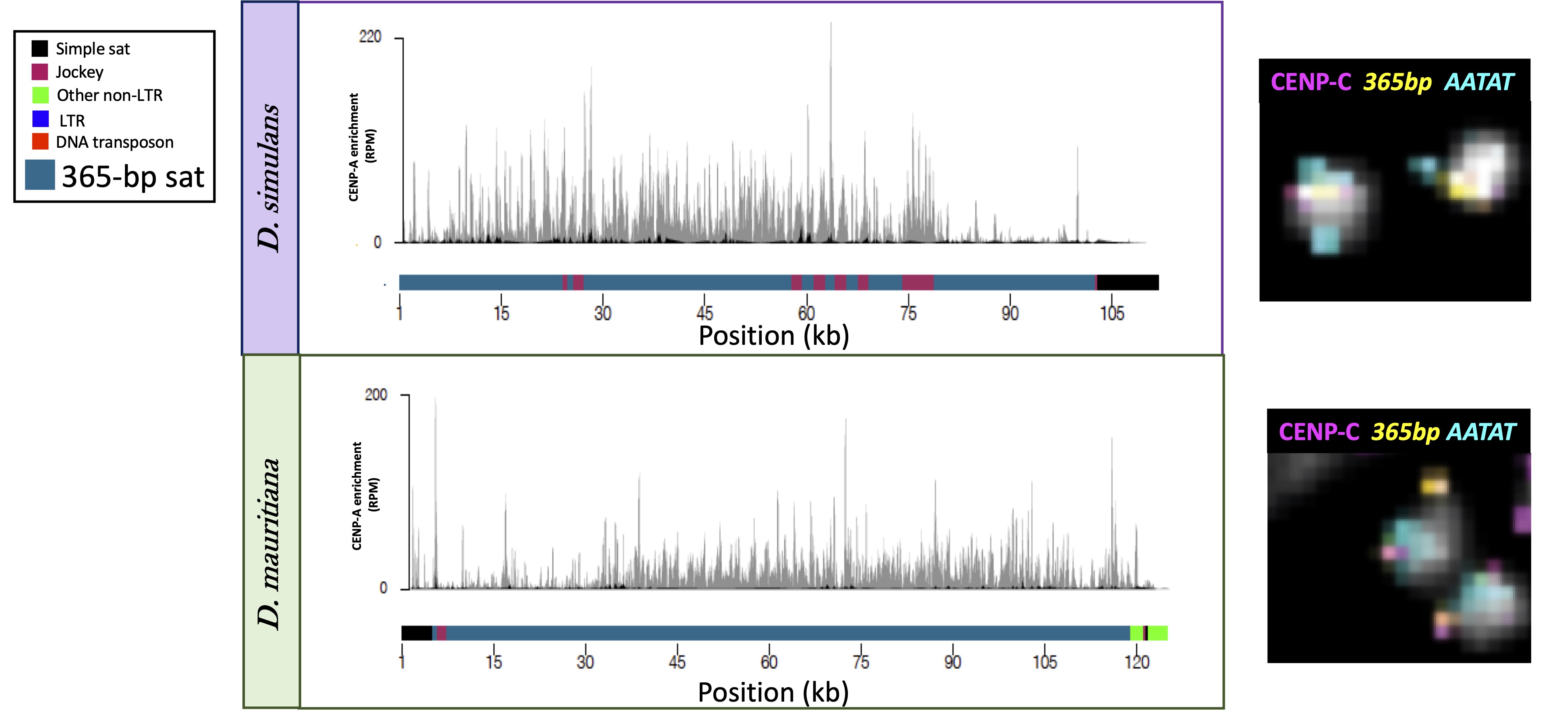
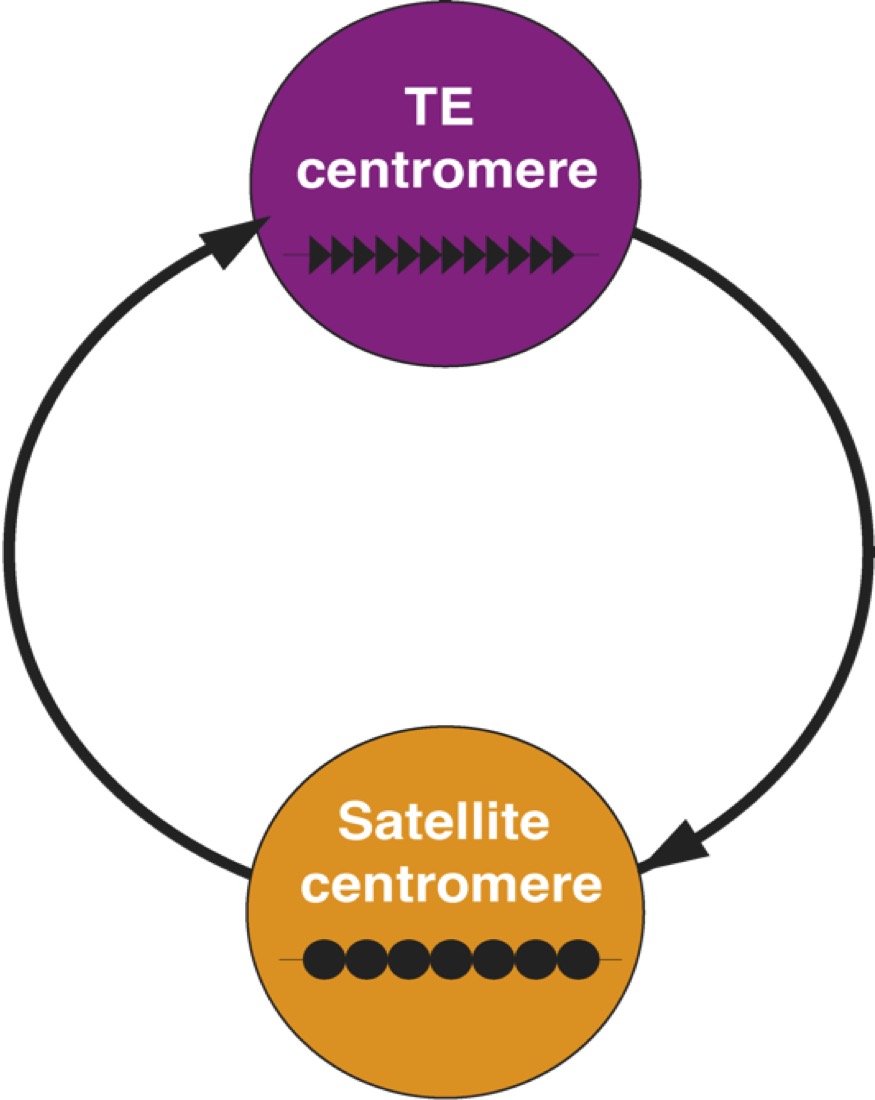
Centromeres are essential chromosomal regions required for proper chromosome segregation during cell division. Disruptions in centromere function can lead to chromosomal instability, one of the hallmarks of cancer. Despite their essential function, centromeres evolve rapidly. In many species, centromeres are located in highly repetitive genome regions but the contribution of DNA sequences to centromere function is unclear, as centromere identity is determined by a centromere-specific histone variant. Drosophila species are fantastic models to study centromere function and evolution: they have small but complex genomes and are powerhouse model organisms for doing genetics. We combine comparative (epi)genomic and cytogenetic approaches to study the evolution of centromere organization in Drosophila species. Together with our collaborators, we discovered that Drosophila melanogaster centromeres correspond to islands of retroelements embedded in satellite DNA. Our current focus is on the role of centromeric DNA in centromere function and the evolutionary forces shaping the rapid evolution. We see dramatic shifts in centromere composition and organization over short evolutionary time periods in Drosophila species. We are testing the hypothesis that genetic conflicts contribute to the rapid evolutionary dynamics at centromeres.